Background knowledge for the Tidy Transcriptomics workshop
Maria Doyle, Peter MacCallum Cancer Centre1
Stefano Mangiola, Walter and Eliza Hall Institute2
Source:vignettes/background.Rmd
background.Rmd
Schedule
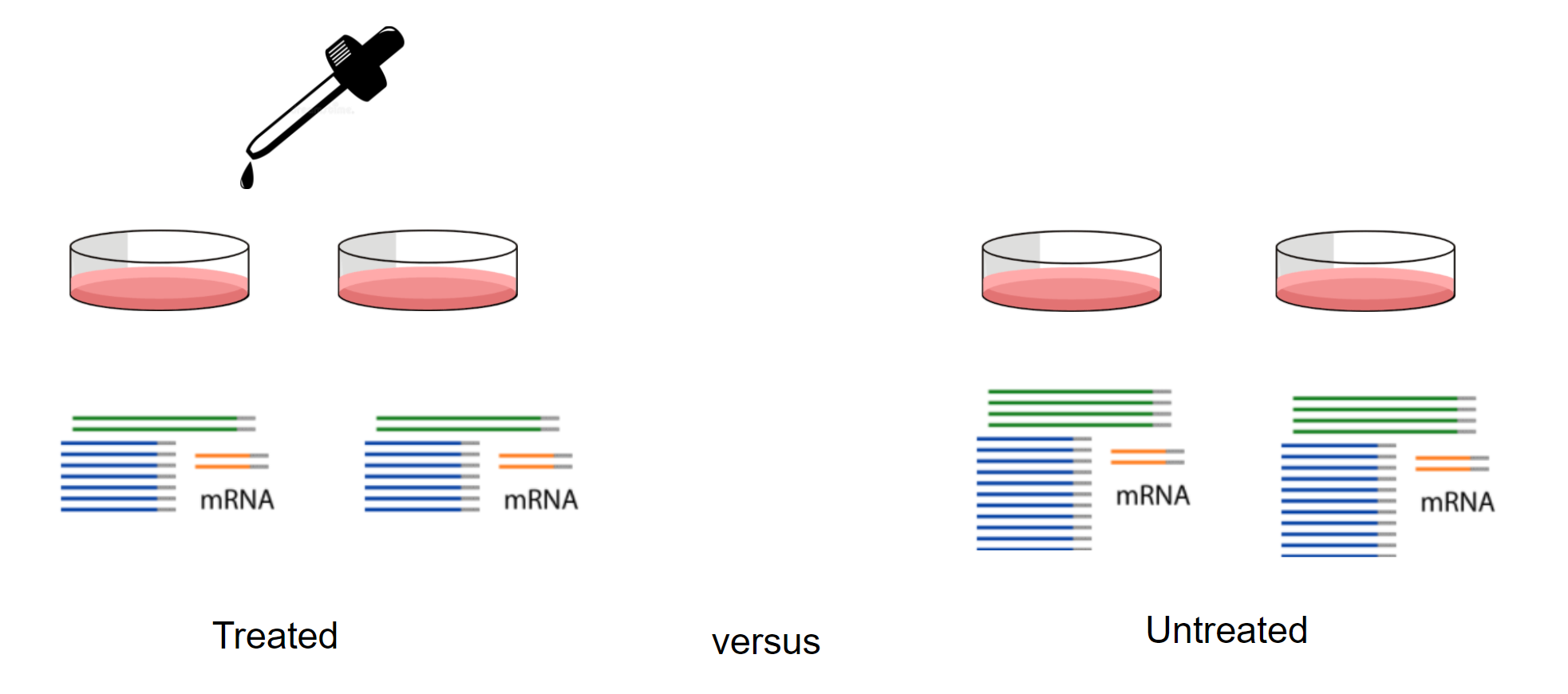
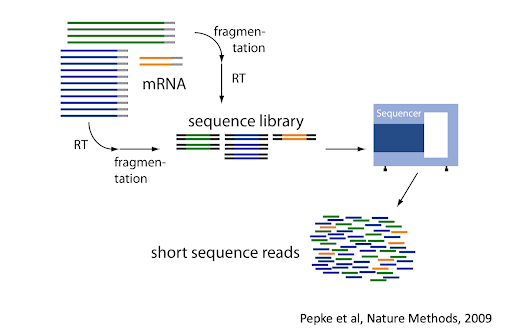
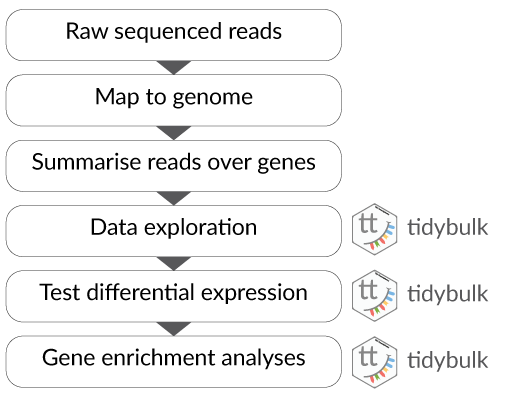
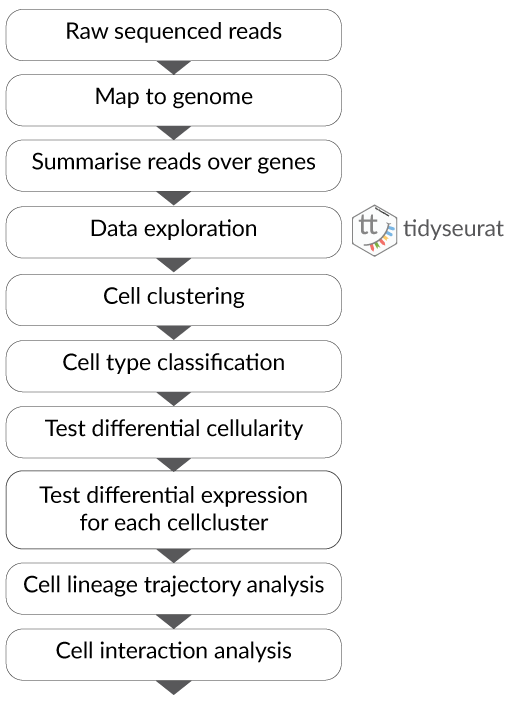
- Part 1 Tidy Bulk RNA-seq analysis
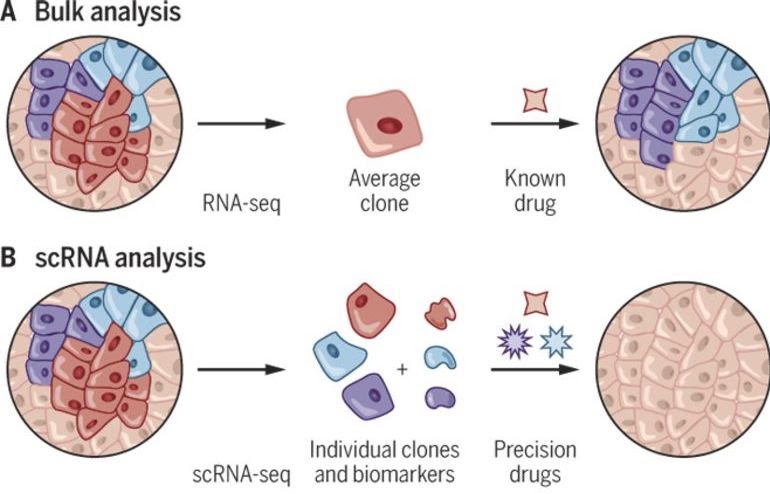
- Part 2 Tidy Single-cell RNA-seq analysis
A more detailed schedule can be found here.
Format: a few introductory slides, then demos, exercises plus Q&A.
Interact: Zoom chat and polls
Poll: Will you code along and/or try out exercises during this workshop?Poll: Do you have experience with transcriptomic analyses?Poll: Do you have experience with tidyverse?Poll: What are you more interested in learning in this workshop?What is transcriptomics?
“The transcriptome is the set of all RNA transcripts, including coding and non-coding, in an individual or a population of cells”
Wikipedia
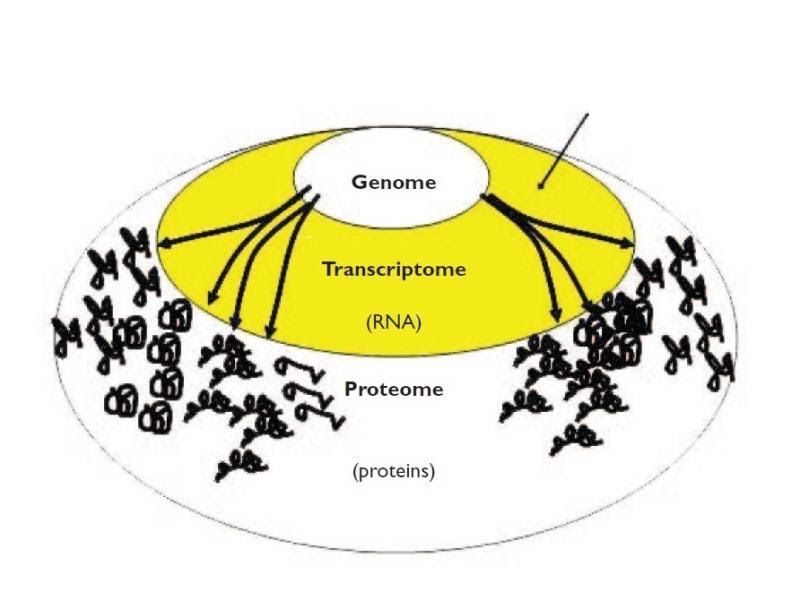
Why use transcriptomics?
- Genome (DNA) pretty stable
- Proteome (proteins) harder to measure
- Transcriptome (RNA) can measure changes in expression of thousands of coding and non-coding transcripts
Types of transcriptomic analyses
-
Differential expression
-
Cell type composition
- Alternative splicing
- Novel transcript discovery
- Fusions identification
- Variant analysis
Topics in bold we will see in this workshop
Getting started
Cloud
Easiest way to run this material. Only available during workshop. Thanks to ARDC for providing RStudio in the Nectar Research Cloud and Andy Botting from ARDC for helping to set up.
- Login to https://rstudio.tidytranscriptomics-workshop.cloud.edu.au/rstudio/auth-sign-in with one of the supplied usernames and passwords
- Open
tidytranscriptomics.Rmdinvignettesfolder
Local
If you want to install on your own computer, see instructions here. We recommend using the Cloud during the ABACBS workshop and this method if you want to run the material after the workshop.