get_heatmap_data() extracts the heatmap matrix as it appears in the plot along with the row and column dendrograms, all with consistent naming.
get_heatmap_data(.data)
# S4 method for class 'InputHeatmap'
get_heatmap_data(.data)Source
[Mangiola and Papenfuss., 2020](https://joss.theoj.org/papers/10.21105/joss.02472)
Value
A list containing:
matrix: The abundance matrix with rows and columns ordered as in the heatmap
row_dend: The row dendrogram object
column_dend: The column dendrogram object
A list containing the ordered matrix, row dendrogram, and column dendrogram
Details
This function converts the InputHeatmap to ComplexHeatmap, draws it to perform clustering, then extracts the ordered matrix and dendrograms exactly as they appear in the heatmap plot.
References
Mangiola, S. and Papenfuss, A.T., 2020. "tidyHeatmap: an R package for modular heatmap production based on tidy principles." Journal of Open Source Software. doi:10.21105/joss.02472.
Examples
hm <- tidyHeatmap::N52 |>
tidyHeatmap::heatmap(
.row = symbol_ct,
.column = UBR,
.value = `read count normalised log`
) |>
annotation_group(
CAPRA_TOTAL,
palette_grouping = list(c("#E64B35", "#4DBBD5")),
group_label_fontsize = 10,
show_group_name = FALSE
)
# Multiple grouping variables
tidyHeatmap::N52 |>
tidyHeatmap::heatmap(
.row = symbol_ct,
.column = UBR,
.value = `read count normalised log`
) |>
annotation_group(
CAPRA_TOTAL,
`Cell type`,
palette_grouping = list(
c("#E64B35", "#4DBBD5"), # colors for CAPRA_TOTAL
c("#00A087", "#F39B7F") # colors for Cell type
)
)
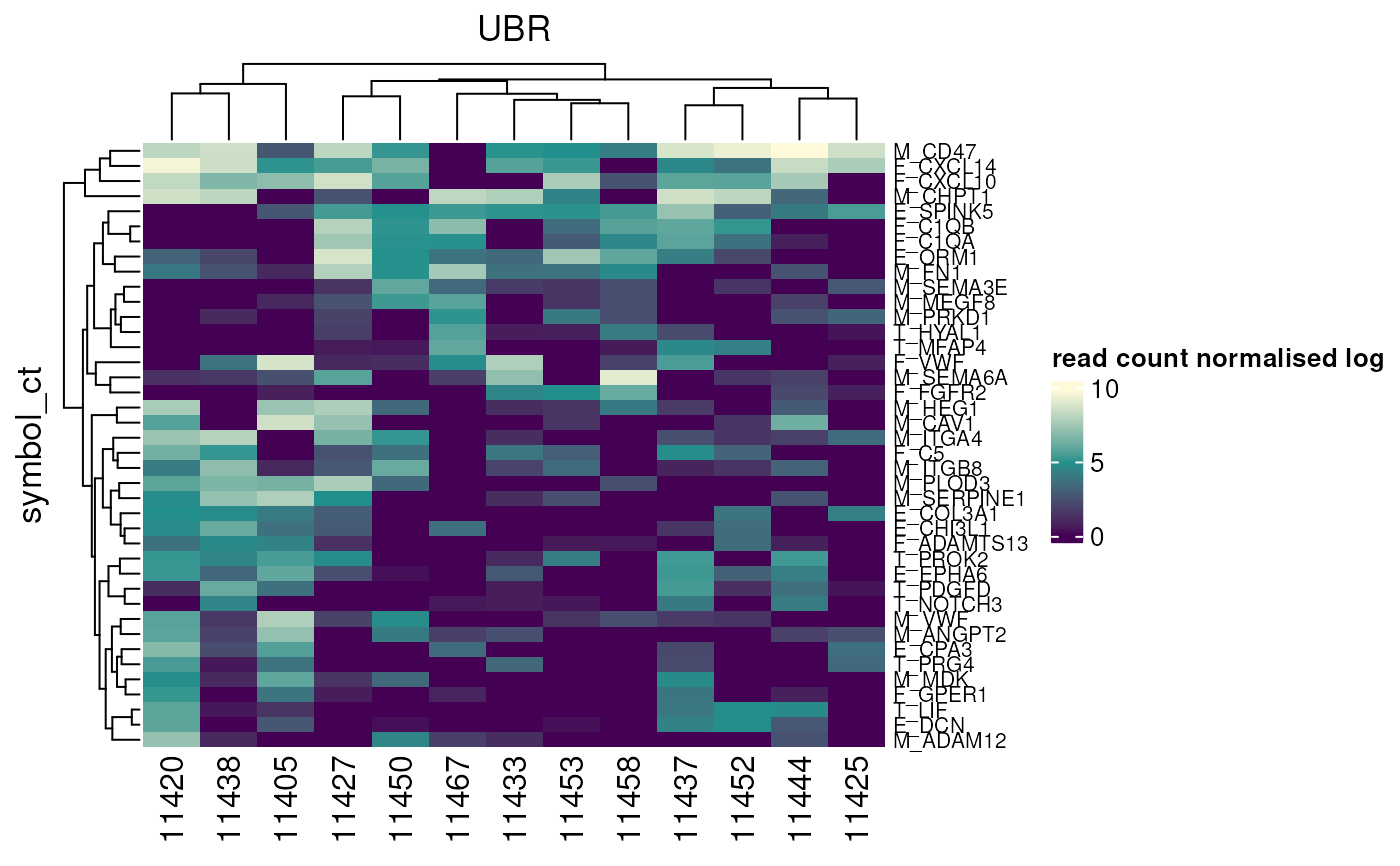 # Get heatmap data as plotted
result <- hm |> get_heatmap_data()
# Get heatmap data as plotted
result <- hm |> get_heatmap_data()
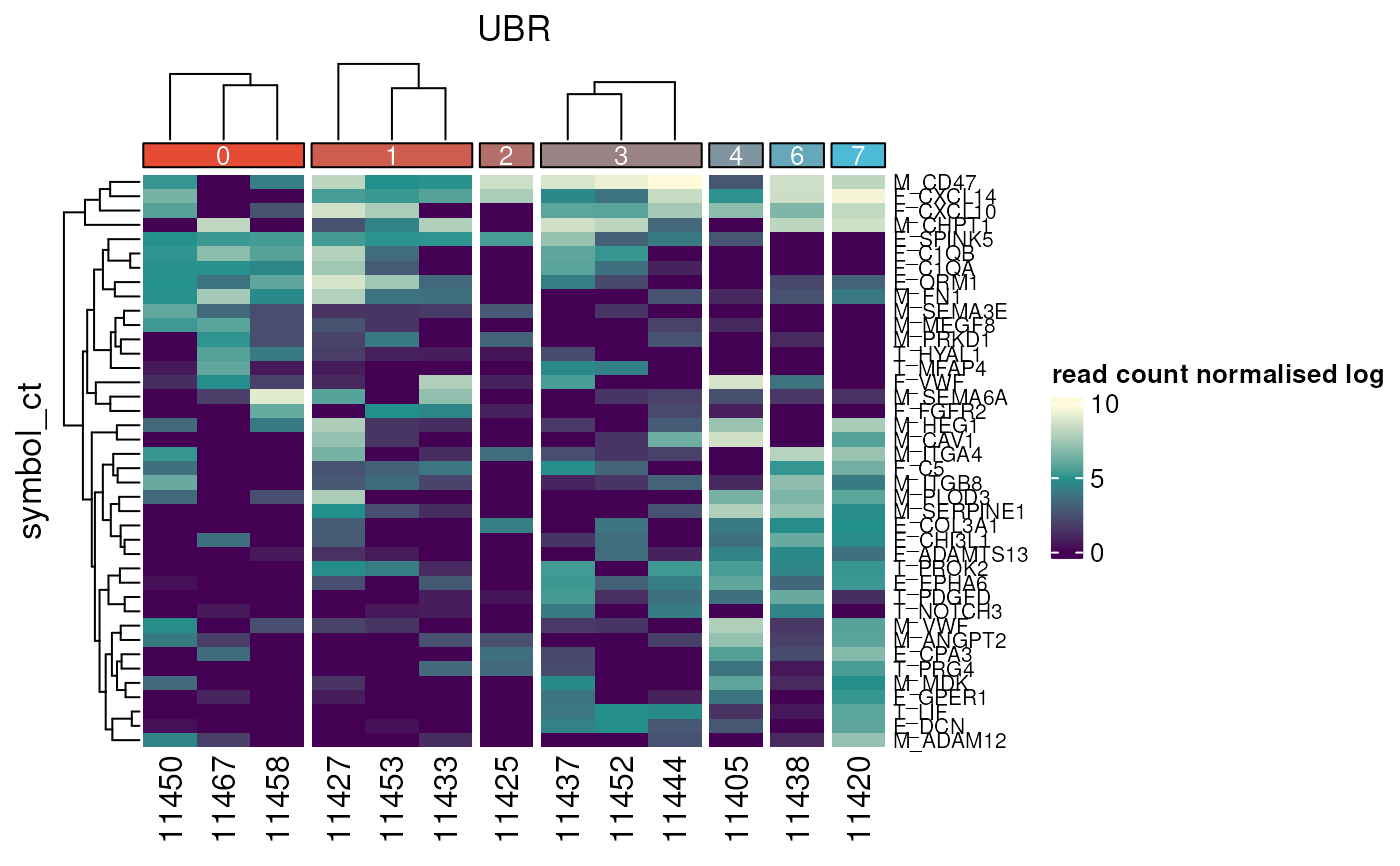 ordered_matrix <- result$matrix
row_dendrogram <- result$row_dend
column_dendrogram <- result$column_dend
ordered_matrix <- result$matrix
row_dendrogram <- result$row_dend
column_dendrogram <- result$column_dend